(For a full list of publications see below or go to Google Scholar)
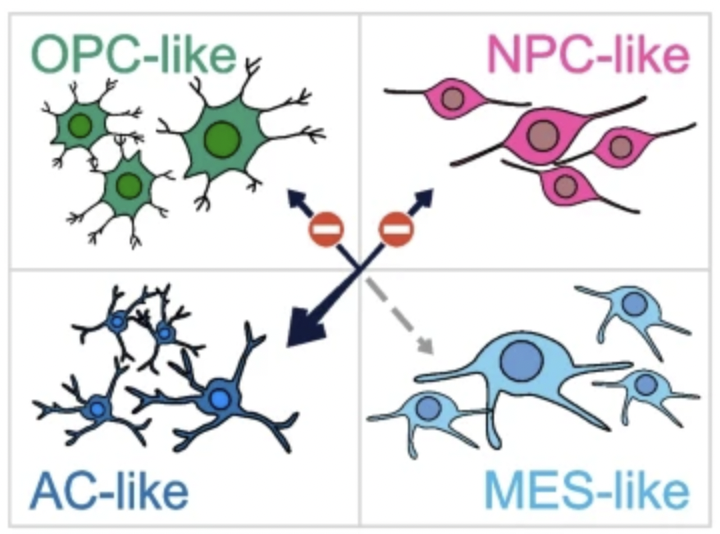
DNA methylation plays a crucial role in cancer development and progression and has been linked to genetically and clinically distinct tumor classes, including IDH-mutated and IDH-wildtype adult-type diffuse gliomas. Here, we identify a CpG-island methylator phenotype (CIMP) that characterizes the receptor tyrosine kinase 2 (RTK2) subtype of IDH-wildtype glioblastoma.
Costa AL, Doncevic D, Wu Y, Yang L, Man KH, Spreng AS, Winter H, Fletcher MNC, Radlwimmer B, Herrmann, C
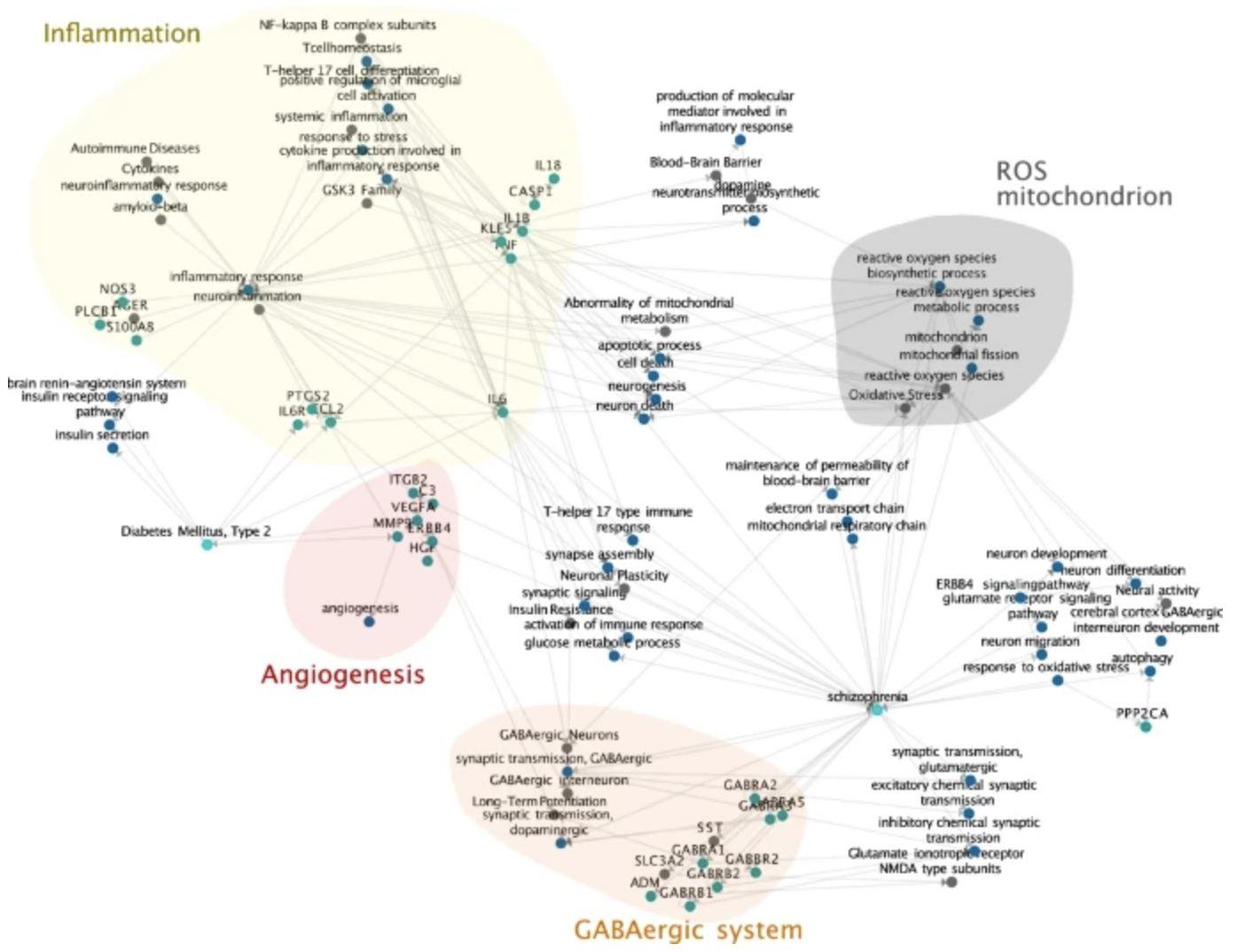
The clinical burden of mental illness, in particular schizophrenia and bipolar disorder, are driven by frequent chronic courses and increased mortality, as well as the risk for comorbid conditions such as cardiovascular disease and type 2 diabetes. Evidence suggests an overlap of molecular pathways between psychotic disorders and somatic comorbidities. In this study, we developed a computational framework to perform comorbidity modeling via an improved integrative unsupervised machine learning approach based on multi-rank non-negative matrix factorization (mrNMF). Using this procedure, we extracted molecular signatures potentially explaining shared comorbidity mechanisms.
Zhang, Y., Bharadhwaj, VS., Kodamullil AT, Herrmann, C.
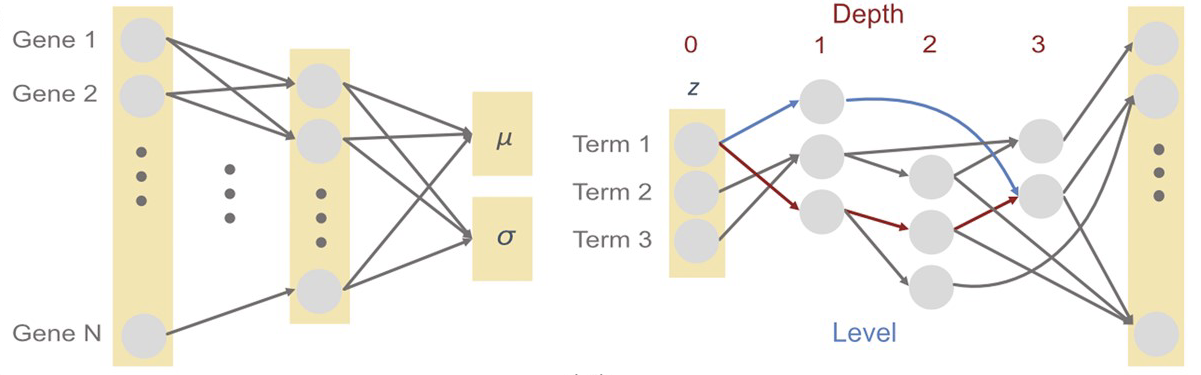
To shed light on the inner workings of VAE and enable direct interpretability of the model through its structure, we designed a novel VAE, OntoVAE (Ontology guided VAE) that can incorporate any ontology in its latent space and decoder part and, thus, provide pathway or phenotype activities for the ontology terms. In this work, we demonstrate that OntoVAE can be applied in the context of predictive modeling and show its ability to predict the effects of genetic or drug-induced perturbations using different ontologies and both, bulk and single-cell transcriptomic datasets. Finally, we provide a flexible framework, which can be easily adapted to any ontology and dataset.
Doncevic, D., Herrmann, C.
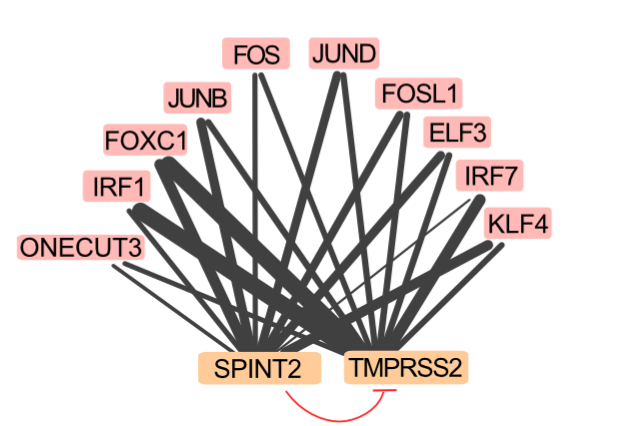
We describe a new important actor which plays a decisive role in the entry of the virus into the cell. The serine protease inhibitor SPINT2 inhibits the well described serine protease TMPRSS2, and we show that inhibiting or overexpressing this gene leads to an increase, respectively a strong decrease of the number of infected cells. Intriguingly, this gene has been described in the context of diseases which are known to be comorbid with COVID19, for example Type 2 diabetes and several malignancies.
Carlos Ramirez Alvarez, Carmon Kee, Ashwini Kumar Sharma, Leonie Thomas, Florian I Schmidt, Megan L Stanifer, Steeve Boulant*, Carl Herrmann*
PLOS Pathog 17(6): e1009687 (2021)
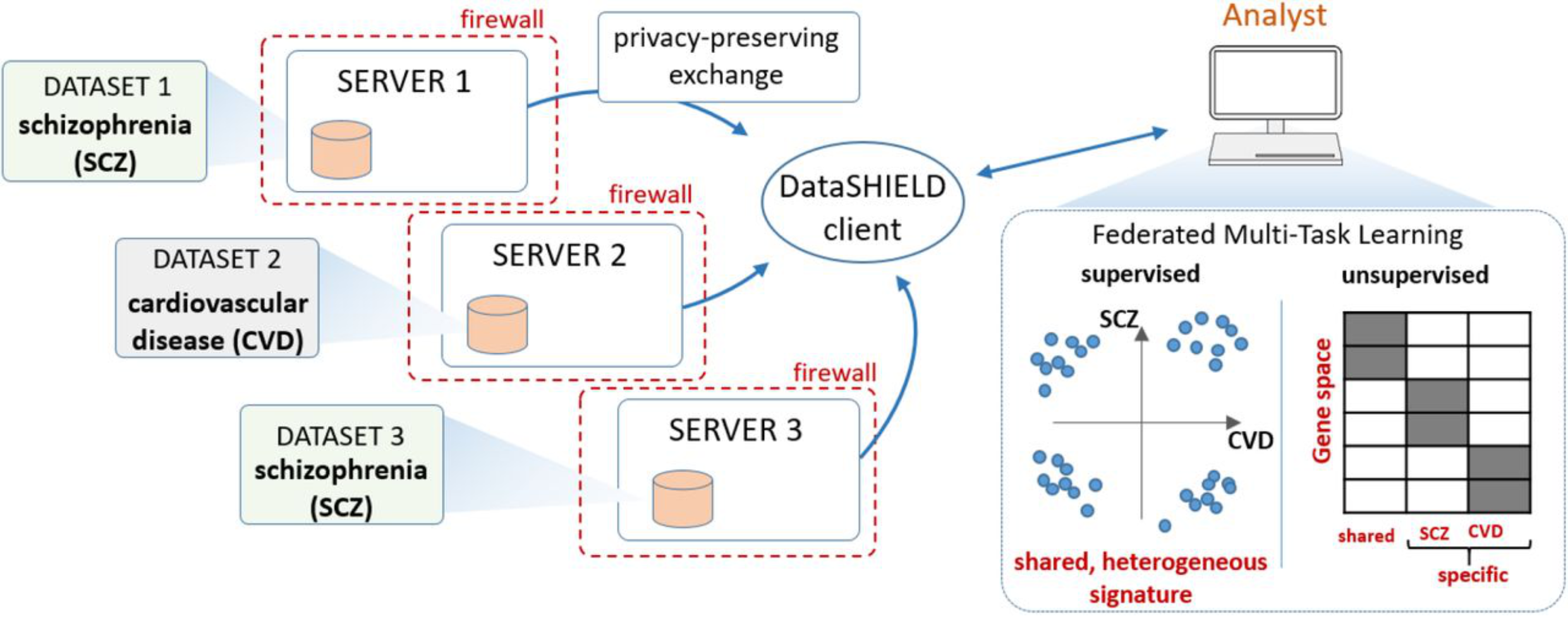
Multitask learning allows the simultaneous learning of multiple algorithms. Here, we describe the development of dsMTL, a computational framework for privacy-preserving, distributed multi-task machine learning that includes three supervised and one unsupervised algorithms.
Han Cao*, Youcheng Zhang*, Jan Baumbach, Paul R Burton, Dominic Dwyer, Nikolaos Koutsouleris, Julian Matschinske, Yannick Marcon, Sivanesan Rajan, Thilo Rieg, Patricia Ryser-Welch, Julian Späth, Carl Herrmann*, Emanuel Schwarz*
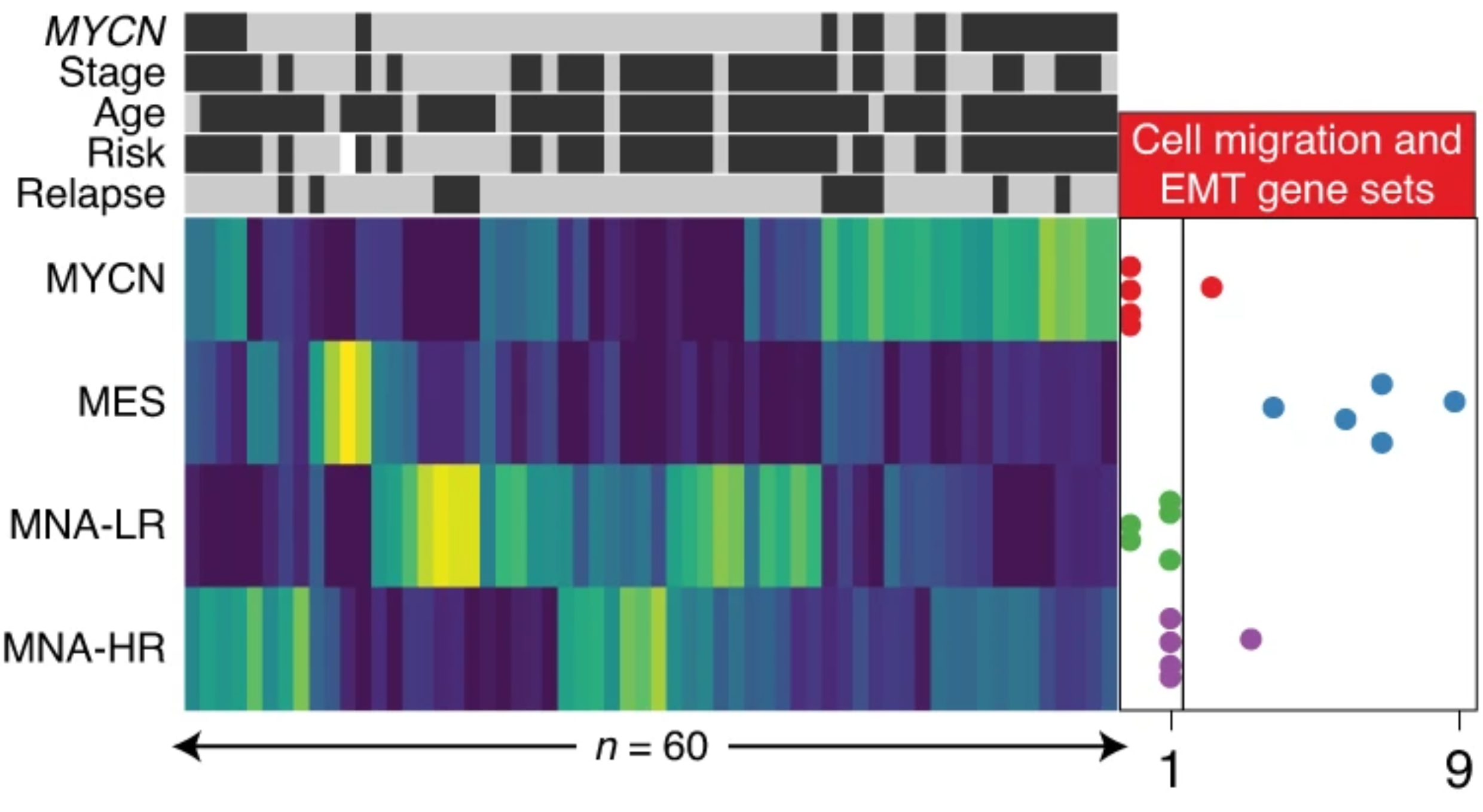
Using genome-wide H3K27ac profiles across 60 NBs, we identified four major super-enhancer-driven epigenetic subtypes and their underlying master regulatory networks. Three of these subtypes recapitulated known clinical groups, while the fourth subtype, exhibiting mesenchymal characteristics, ressemblance to Schwann cell precursors, was induced by RAS activation and was enriched in relapsed disease. This study reveals subtype-specific super-enhancer regulation in NBs.
Moritz Gartlgruber*, Ashwini Kumar Sharma*, Andrés Quintero*, Daniel Dreidax*, Selina Jansky, Young-Gyu Park, Sina Kreth, Johanna Meder, Daria Doncevic, Paul Saary, Umut H Toprak, Naveed Ishaque, Elena Afanasyeva, Elisa Wecht, Jan Koster, Rogier Versteeg, Thomas GP Grünewald, David TW Jones, Stefan M Pfister, Kai-Oliver Henrich, Johan van Nes, Carl Herrmann*, Frank Westermann*
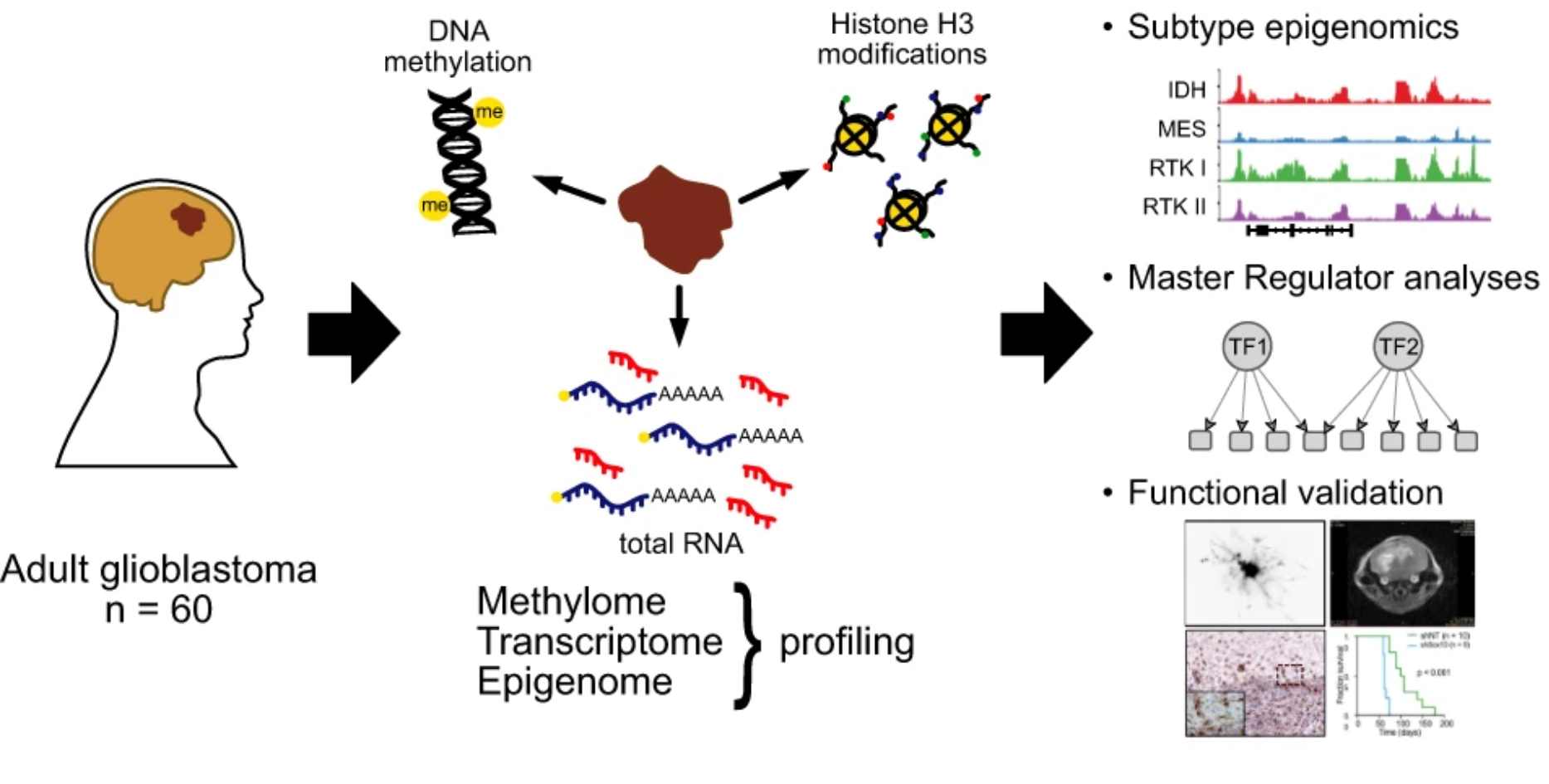
Glioblastoma frequently exhibits therapy-associated subtype transitions to mesenchymal phenotypes with adverse prognosis. Here, we perform multi-omic profiling of 60 glioblastoma primary tumours and identify 38 subtype master regulators. SOX10 repression in an in vivo glioblastoma mouse model results in increased tumour invasion, immune cell infiltration and significantly reduced survival, reminiscent of progressive human glioblastoma. These results identify SOX10 as a bona fide master regulator of the RTK I subtype, with both tumour cell-intrinsic and microenvironmental effects.
Yonghe Wu*, Michael Fletcher*, Zuguang Gu*, Qi Wang, Barbara Costa, Anna Bertoni, Ka-Hou Man, Magdalena Schlotter, Jörg Felsberg, Jasmin Mangei, Martje Barbus, Ann-Christin Gaupel, Wei Wang, Tobias Weiss, Roland Eils, Michael Weller, Haikun Liu, Guido Reifenberger, Andrey Korshunov, Peter Angel, Peter Lichter, Carl Herrmann*, Bernhard Radlwimmer*

Non-negative matrix factorization (NMF) has been widely used for the analysis of genomic data to perform feature extraction and signature identification due to the interpretability of the decomposed signatures. However, running a basic NMF analysis requires the installation of multiple tools and dependencies, along with a steep learning curve and computing time. To mitigate such obstacles, we developed ShinyButchR, a novel R/Shiny application that provides a complete NMF-based analysis workflow, allowing the user to perform matrix decomposition using NMF, feature extraction, interactive visualization, relevant signature identification, and association to biological and clinical variables. ShinyButchR builds upon the also novel R package ButchR, which provides new TensorFlow solvers for algorithms of the NMF family, functions for downstream analysis, a rational method to determine the optimal factorization rank and a novel feature selection strategy.
Andres Quintero, Daniel Hübschmann, Nils Kurzawa, Sebastian Steinhauser, Philipp Rentzsch, Stephen Krämer, Carolin Andresen, Jeongbin Park, Roland Eils, Matthias Schlesner, Carl Herrmann
Biology Methods and Protocols (2020)
A new IDH-independent hypermethylation phenotype is associated with astrocyte-like cell state in glioblastoma
Costa AL, Doncevic D, Wu Y, Yang L, Man KH, Spreng AS, Winter H, Fletcher MNC, Radlwimmer B, Herrmann, C
Genome Biology (2025)
A network of transcriptomic signatures identifies novel comorbidity mechanisms between schizophrenia and somatic disorders
Zhang, Y., Bharadhwaj, VS., Kodamullil AT, Herrmann, C.
Discover Mental Health (2024)
Interleukin-22 Promotes Cell Proliferation to Combat Virus Infection in Human Intestinal Epithelial Cells
Guo, C., Sharma, AK., Guzman, J., Herrmann, C., Boulant, S., Stanifer, M.
Journal of Interferon and Cytokine Research (2024)
Biologically informed variational autoencoders allow predictive modeling of genetic and drug-induced perturbations
Doncevic, D., Herrmann, C.
Bioinformatics (2023)
Genomic profiling of HIV-1 integration in microglia cells links viral integration to the topologically associated domains
Rheinberger, M., Costa, A.L., Kampmann, M., Glavas, D., Shytaj, I.L., Sreeram, S., Penzo, C., Tibroni, N., Garcia-Mesa, Y., Leskov, K., Fackler, O.T., Vlahovicek, K., Karn, J., Lucic, B., Herrmann, C.*, Lusic, M.*
Cell Reports (2023)
Exploring the impact of dexamethasone on gene regulation in myeloma cells
Victor Gaborit, Jonathan Cruard, Catherine Guérin-Charbonnel, Jennifer Derrien, Jean-Baptiste Alberge, Elise Douillard, Nathalie Roi, Magali Devic, Loïc Campion, Frank Westermann, Philippe Moreau, Carl Herrmann, Jérémie Bourdon, Florence Magrangeas, Stephane Minvielle
Life Science Alliance (2023)
dsMTL: a computational framework for privacy-preserving, distributed multi-task machine learning
Cao, H., Zhang, Y., Baumbach, J., Burton, P.R., Dwyer, D., Koutsouleris, N., Matschinske, J., Marcon, Y., Rajan, S., Rieg, T., Ryser-Welch, P., Späth, J., The COMMITMENT Consortium, Herrmann, C.*, Schwarz, E.*
Bioinformatics (2022)
The DNA methylation landscape of multiple myeloma shows extensive inter-and intrapatient heterogeneity that fuels transcriptomic variability
Jennifer Derrien, Catherine Guérin-Charbonnel, Victor Gaborit, Loïc Campion, Magali Devic, Elise Douillard, Nathalie Roi, Hervé Avet-Loiseau, Olivier Decaux, Thierry Facon, Jan-Philipp Mallm, Roland Eils, Nikhil C Munshi, Philippe Moreau, Carl Herrmann, Florence Magrangeas, Stéphane Minvielle
Genome Medicine (2021)
Derepression of retroelements in acute myeloid leukemia with 3q aberrations
Jagoda Mika, Sophie Ottema, Sandra Kiehlmeier, Sabrina Kruse, Leonie Smeenk, Judith Müller, Sabrina Schweiggert, Carl Herrmann, Mathijs Sanders, Ruud Delwel, Stefan Gröschel
Haematologica (2021)
The endogenous cellular protease inhibitor SPINT2 controls SARS-CoV-2 viral infection and is associated to disease severity
Carlos Ramirez Alvarez, Carmon Kee, Ashwini Kumar Sharma, Leonie Thomas, Florian I Schmidt, Megan L Stanifer, Steeve Boulant*, Carl Herrmann*
PLOS Pathog 17(6): e1009687 (2021)
Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma
Selina Jansky, Ashwini Kumar Sharma, Verena Körber, Andrés Quintero, Umut H Toprak, Elisa M Wecht, Moritz Gartlgruber, Alessandro Greco, Elad Chomsky, Thomas GP Grünewald, Kai-Oliver Henrich, Amos Tanay, Carl Herrmann, Thomas Höfer, Frank Westermann
Nature genetics (2021)
Kalirin-RAC controls nucleokinetic migration in ADRN-type neuroblastoma
Elena A Afanasyeva, Moritz Gartlgruber, Tatsiana Ryl, Bieke Decaesteker, Geertrui Denecker, Gregor Mönke, Umut H Toprak, Andres Florez, Alica Torkov, Daniel Dreidax, Carl Herrmann, Konstantin Okonechnikov, Sara Ek, Ashwini Kumar Sharma, Vitaliya Sagulenko, Frank Speleman, Kai-Oliver Henrich, Frank Westermann
Life science alliance (2021)
Identification of therapeutic targets of the hijacked super-enhancer complex in EVI1-rearranged leukemia
Sandra Kiehlmeier, Mahmoud-Reza Rafiee, Ali Bakr, Jagoda Mika, Sabrina Kruse, Judith Müller, Sabrina Schweiggert, Carl Herrmann, Gianluca Sigismondo, Peter Schmezer, Jeroen Krijgsveld, Stefan Gröschel
Leukemia (2021)
Single‐cell analyses reveal SARS‐CoV‐2 interference with intrinsic immune response in the human gut
Sergio Triana, Camila Metz‐Zumaran, Carlos Ramirez, Carmon Kee, Patricio Doldan, Mohammed Shahraz, Daniel Schraivogel, Andreas R Gschwind, Ashwini K Sharma, Lars M Steinmetz, Carl Herrmann, Theodore Alexandrov, Steeve Boulant, Megan L Stanifer
Molecular systems biology (2021)
Identifying multimodal signatures underlying the somatic comorbidity of psychosis: the COMMITMENT roadmap
Emanuel Schwarz, Dag Alnæs, Ole A Andreassen, Han Cao, Junfang Chen, Franziska Degenhardt, Daria Doncevic, Dominic Dwyer, Roland Eils, Jeanette Erdmann, Carl Herrmann, Martin Hofmann-Apitius, Tobias Kaufmann, Nikolaos Koutsouleris, Alpha T Kodamullil, Adyasha Khuntia, Sören Mucha, Markus M Nöthen, Riya Paul, Mads L Pedersen, Andres Quintero, Heribert Schunkert, Ashwini Sharma, Heike Tost, Lars T Westlye, Youcheng Zhang, Andreas Meyer-Lindenberg
Molecular Psychiatry (2021)
Alternative lengthening of telomeres in childhood neuroblastoma from genome to proteome
Sabine A Hartlieb, Lina Sieverling, Michal Nadler-Holly, Matthias Ziehm, Umut H Toprak, Carl Herrmann, Naveed Ishaque, Konstantin Okonechnikov, Moritz Gartlgruber, Young-Gyu Park, Elisa Maria Wecht, Larissa Savelyeva, Kai-Oliver Henrich, Carolina Rosswog, Matthias Fischer, Barbara Hero, David TW Jones, Elke Pfaff, Olaf Witt, Stefan M Pfister, Richard Volckmann, Jan Koster, Katharina Kiesel, Karsten Rippe, Sabine Taschner-Mandl, Peter Ambros, Benedikt Brors, Matthias Selbach, Lars Feuerbach, Frank Westermann
Nature communications (20201)
dsMTL-a computational framework for privacy-preserving, distributed multi-task machine learning
Han Cao*, Youcheng Zhang*, Jan Baumbach, Paul R Burton, Dominic Dwyer, Nikolaos Koutsouleris, Julian Matschinske, Yannick Marcon, Sivanesan Rajan, Thilo Rieg, Patricia Ryser-Welch, Julian Späth, Carl Herrmann*, Emanuel Schwarz*
Bioinformatics (2022)
MYCN mediates cysteine addiction and sensitizes to ferroptosis
Hamed Alborzinia, Andres F Florez, Sina Gogolin, Lena M Brueckner, Chunxuan Shao, Moritz Gartlgruber, Michal Nadler Holly, Matthias Ziehm, Franziska Paul, Sebastian Steinhauser, Emma Bell, Marjan Shaikhkarami, Sabine Hartlieb, Daniel Dreidax, Elisa M Hess, Jochen Kreth, Gernot Poschet, Michael Buettner, Barbara Nicke, Carlo Stresemann, Jan H Reiling, Matthias Fischer, Ido Amit, Matthias Selbach, Carl Herrmann, Stefan Woelfl, Kai Oliver Henrich, Thomas Hoefer, Frank Westermann
bioRxiv (2021)
Integrative ranking of enhancer networks facilitates the discovery of epigenetic markers in cancer
Qi Wang, Yonghe Wu, Tim Vorberg, Roland Eils, Carl Herrmann
Frontiers in genetics (2021)
Super enhancers define regulatory subtypes and cell identity in neuroblastoma
Moritz Gartlgruber*, Ashwini Kumar Sharma*, Andrés Quintero*, Daniel Dreidax*, Selina Jansky, Young-Gyu Park, Sina Kreth, Johanna Meder, Daria Doncevic, Paul Saary, Umut H Toprak, Naveed Ishaque, Elena Afanasyeva, Elisa Wecht, Jan Koster, Rogier Versteeg, Thomas GP Grünewald, David TW Jones, Stefan M Pfister, Kai-Oliver Henrich, Johan van Nes, Carl Herrmann*, Frank Westermann*
Nature Cancer (2021)
Glioblastoma epigenome profiling identifies SOX10 as a master regulator of molecular tumour subtype
Yonghe Wu*, Michael Fletcher*, Zuguang Gu*, Qi Wang, Barbara Costa, Anna Bertoni, Ka-Hou Man, Magdalena Schlotter, Jörg Felsberg, Jasmin Mangei, Martje Barbus, Ann-Christin Gaupel, Wei Wang, Tobias Weiss, Roland Eils, Michael Weller, Haikun Liu, Guido Reifenberger, Andrey Korshunov, Peter Angel, Peter Lichter, Carl Herrmann*, Bernhard Radlwimmer*
Nature communications (2020)
Quantitative proteomics reveals specific metabolic features of Acute Myeloid Leukemia stem cells
Simon Raffel, Daniel Klimmeck, Mattia Falcone, Aykut Demir, Alireza Pouya, Petra Zeisberger, Christoph Lutz, Marco Tinelli, Oliver Bischel, Lars Bullinger, Christian Thiede, Anne Flörcken, Jörg Westermann, Gerhard Ehninger, Anthony D Ho, Carsten Müller-Tidow, Zuguang Gu, Carl Herrmann, Jeroen Krijgsveld, Andreas Trumpp, Jenny Hansson
Blood (2020)
Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples
Matthew H. Bailey, William U. Meyerson, Lewis Jonathan Dursi, Liang-Bo Wang, Guanlan Dong, Wen-Wei Liang, Amila Weerasinghe, Shantao Li, Yize Li, Sean Kelso, MC3 Working Group, PCAWG novel somatic mutation calling methods working group, Gordon Saksena, Kyle Ellrott, Michael C. Wendl, David A. Wheeler, Gad Getz, Jared T. Simpson, Mark B. Gerstein, Li Ding & PCAWG Consortium
Nature Communications (2020)
Sex differences in oncogenic mutational processes
Constance H. Li, Stephenie D. Prokopec, Ren X. Sun, Fouad Yousif, Nathaniel Schmitz, PCAWG Tumour Subtypes and Clinical Translation, Paul C. Boutros & PCAWG Consortium
Nature Communications (2020)
Automated 3D light-sheet screening with high spatiotemporal resolution reveals mitotic phenotypes
Björn Eismann, Teresa G Krieger, Jürgen Beneke, Ruben Bulkescher, Lukas Adam, Holger Erfle, Carl Herrmann, Roland Eils, Christian Conrad
Journal of cell science (2020)
Passenger mutations in more than 2,500 cancer genomes: overall molecular functional impact and consequences
Sushant Kumar, Jonathan Warrell, Shantao Li, Patrick D McGillivray, William Meyerson, Leonidas Salichos, Arif Harmanci, Alexander Martinez-Fundichely, Calvin WY Chan, Morten Muhlig Nielsen, Lucas Lochovsky, Yan Zhang, Xiaotong Li, Shaoke Lou, Jakob Skou Pedersen, Carl Herrmann, Gad Getz, Ekta Khurana, Mark B Gerstein
Cell (2020)
Combined burden and functional impact tests for cancer driver discovery using DriverPower
Shimin Shuai, PCAWG Drivers and Functional Interpretation Working Group, Steven Gallinger, Lincoln Stein & PCAWG Consortium
Nature Communications (2021)
Analyses of non-coding somatic drivers in 2,658 cancer whole genomes
Esther Rheinbay, Morten Muhlig Nielsen,[…] PCAWG Consortium
Nature (2020)
Pan-cancer analysis of whole genomes
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium
Nature (2020)
ShinyButchR: Interactive NMF-based decomposition workflow of genome-scale datasets
Andres Quintero, Daniel Hübschmann, Nils Kurzawa, Sebastian Steinhauser, Philipp Rentzsch, Stephen Krämer, Carolin Andresen, Jeongbin Park, Roland Eils, Matthias Schlesner, Carl Herrmann
Biology Methods and Protocols (2020)
Single-nucleus chromatin accessibility reveals intratumoral epigenetic heterogeneity in IDH1 mutant gliomas
Ruslan Al-Ali, Katharina Bauer, Jong-Whi Park, Ruba Al Abdulla, Valentina Fermi, Andreas von Deimling, Christel Herold-Mende, Jan-Philipp Mallm, Carl Herrmann, Wolfgang Wick, Şevin Turcan
Acta neuropathologica communications (2019)
Impact of cancer mutational signatures on transcription factor motifs in the human genome
Calvin Wing Yiu Chan, Zuguang Gu, Matthias Bieg, Roland Eils, Carl Herrmann
BMC medical genomics (2019)
Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity
Longqi Liu, Chuanyu Liu, Andrés Quintero, Liang Wu, Yue Yuan, Mingyue Wang, Mengnan Cheng, Lizhi Leng, Liqin Xu, Guoyi Dong, Rui Li, Yang Liu, Xiaoyu Wei, Jiangshan Xu, Xiaowei Chen, Haorong Lu, Dongsheng Chen, Quanlei Wang, Qing Zhou, Xinxin Lin, Guibo Li, Shiping Liu, Qi Wang, Hongru Wang, J Lynn Fink, Zhengliang Gao, Xin Liu, Yong Hou, Shida Zhu, Huanming Yang, Yunming Ye, Ge Lin, Fang Chen, Carl Herrmann, Roland Eils, Zhouchun Shang, Xun Xu
Nature communications (2019)
Identification of embryonic neural plate border stem cells and their generation by direct reprogramming from adult human blood cells
Marc Christian Thier, Oliver Hommerding, Jasper Panten, Roberta Pinna, Diego García-González, Thomas Berger, Philipp Wörsdörfer, Yassen Assenov, Roberta Scognamiglio, Adriana Przybylla, Paul Kaschutnig, Lisa Becker, Michael D Milsom, Anna Jauch, Jochen Utikal, Carl Herrmann, Hannah Monyer, Frank Edenhofer, Andreas Trumpp
Cell stem cell (2019)
Induced aneuploidy disrupts MCF10A acini formation and CCND1 expression
Marcel Waschow, Qi Wang, Paul Saary, Corinna Klein, Sabine Aschenbrenner, Katharina Jechow, Lorenz Maier, Stephan Tirier, Brigitte Schoell, Ilse Chudoba, Christian Dietz, Daniel Dreidax, Anna Jauch, Martin Sprick, Carl Herrmann, Roland Eils, Christian Conrad
bioRxiv (2019)
TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets
Bieke Decaesteker, Geertrui Denecker, Christophe Van Neste, Emmy M Dolman, Wouter Van Loocke, Moritz Gartlgruber, Carolina Nunes, Fanny De Vloed, Pauline Depuydt, Karen Verboom, Dries Rombaut, Siebe Loontiens, Jolien De Wyn, Waleed M Kholosy, Bianca Koopmans, Anke HW Essing, Carl Herrmann, Daniel Dreidax, Kaat Durinck, Dieter Deforce, Filip Van Nieuwerburgh, Anton Henssen, Rogier Versteeg, Valentina Boeva, Gudrun Schleiermacher, Johan van Nes, Pieter Mestdagh, Suzanne Vanhauwaert, Johannes H Schulte, Frank Westermann, Jan J Molenaar, Katleen De Preter, Frank Speleman
Nature communications (2018)
Development of early childhood asthma goes along with massive enhancer activation in blood cells
S Trump, M Kloes, M Schmidt, T Bauer, N Ishaque, L Thurmann, M Bieg, C Herrmann, S Roeder, M Bauer, D Weichenhan, O Muecke, C Plass, M Borte, E Von Mutius, M Kabesch, G Stangl, R Lauener, J Pekkanen, J Dalphin, J Riedler, R Eils, I Lehmann
ALLERGY (2018)
BCAT1 restricts αKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation
Simon Raffel, Mattia Falcone, Niclas Kneisel, Jenny Hansson, Wei Wang, Christoph Lutz, Lars Bullinger, Gernot Poschet, Yannic Nonnenmacher, Andrea Barnert, Carsten Bahr, Petra Zeisberger, Adriana Przybylla, Markus Sohn, Martje Tönjes, Ayelet Erez, Lital Adler, Patrizia Jensen, Claudia Scholl, Stefan Fröhling, Sibylle Cocciardi, Patrick Wuchter, Christian Thiede, Anne Flörcken, Jörg Westermann, Gerhard Ehninger, Peter Lichter, Karsten Hiller, Rüdiger Hell, Carl Herrmann, Anthony D Ho, Jeroen Krijgsveld, Bernhard Radlwimmer, Andreas Trumpp
Nature (2017)
Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues
Michael Delacher, Charles D Imbusch, Dieter Weichenhan, Achim Breiling, Agnes Hotz-Wagenblatt, Ulrike Traeger, Ann-Cathrin Hofer, Danny Kägebein, Qi Wang, Felix Frauhammer, Jan-Philipp Mallm, Katharina Bauer, Carl Herrmann, Philipp A Lang, Benedikt Brors, Christoph Plass, Markus Feuerer
Nature immunology (2017)
Identifying personal DNA methylation profiles by genotype inference
Michael Backes, Pascal Berrang, Matthias Bieg, Roland Eils, Carl Herrmann, Mathias Humbert, Irina Lehmann
2017 IEEE Symposium on Security and Privacy (SP) (2017)
Deciphering programs of transcriptional regulation by combined deconvolution of multiple omics layers
Daniel Hüebschmann, Nils Kurzawa, Sebastian Steinhauser, Philipp Rentzsch, Stephen Krämer, Carolin Andresen, Jeongbin Park, Roland Eils, Matthias Schlesner, Carl Herrmann
bioRxiv (2017)
A comprehensive comparison of tools for differential ChIP-seq analysis
Sebastian Steinhauser, Nils Kurzawa, Roland Eils, Carl Herrmann
Briefings in bioinformatics (2016)
Integrative genome-scale analysis identifies epigenetic mechanisms of transcriptional deregulation in unfavorable neuroblastomas
Kai-Oliver Henrich, Sebastian Bender, Maral Saadati, Daniel Dreidax, Moritz Gartlgruber, Chunxuan Shao, Carl Herrmann, Manuel Wiesenfarth, Martha Parzonka, Lea Wehrmann, Matthias Fischer, David J Duffy, Emma Bell, Alica Torkov, Peter Schmezer, Christoph Plass, Thomas Höfer, Axel Benner, Stefan M Pfister, Frank Westermann
Cancer research (2016)
Prenatal maternal stress and wheeze in children: novel insights into epigenetic regulation
Saskia Trump, Matthias Bieg, Zuguang Gu, Loreen Thürmann, Tobias Bauer, Mario Bauer, Naveed Ishaque, Stefan Röder, Lei Gu, Gunda Herberth, Christian Lawerenz, Michael Borte, Matthias Schlesner, Christoph Plass, Nicolle Diessl, Markus Eszlinger, Oliver Mücke, Horst-Dietrich Elvers, Dirk K Wissenbach, Martin Von Bergen, Carl Herrmann, Dieter Weichenhan, Rosalind J Wright, Irina Lehmann, Roland Eils
Scientific reports (2016)
LedPred: an R/bioconductor package to predict regulatory sequences using support vector machines
Denis Seyres, Elodie Darbo, Laurent Perrin, Carl Herrmann, Aitor González
Bioinformatics (2016)
Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes
Pascal D Johann, Serap Erkek, Marc Zapatka, Kornelius Kerl, Ivo Buchhalter, Volker Hovestadt, David TW Jones, Dominik Sturm, Carl Hermann, Maia Segura Wang, Andrey Korshunov, Marina Rhyzova, Susanne Gröbner, Sebastian Brabetz, Lukas Chavez, Susanne Bens, Stefan Gröschel, Fabian Kratochwil, Andrea Wittmann, Laura Sieber, Christina Geörg, Stefan Wolf, Katja Beck, Florian Oyen, David Capper, Peter van Sluis, Richard Volckmann, Jan Koster, Rogier Versteeg, Andreas von Deimling, Till Milde, Olaf Witt, Andreas E Kulozik, Martin Ebinger, Tarek Shalaby, Michael Grotzer, David Sumerauer, Josef Zamecnik, Jaume Mora, Nada Jabado, Michael D Taylor, Annie Huang, Eleonora Aronica, Anna Bertoni, Bernhard Radlwimmer, Torsten Pietsch, Ulrich Schüller, Reinhard Schneppenheim, Paul A Northcott, Jan O Korbel, Reiner Siebert, Michael C Frühwald, Peter Lichter, Roland Eils, Amar Gajjar, Martin Hasselblatt, Stefan M Pfister, Marcel Kool
Cancer cell (2016)
Environment‐induced epigenetic reprogramming in genomic regulatory elements in smoking mothers and their children
Tobias Bauer, Saskia Trump, Naveed Ishaque, Loreen Thürmann, Lei Gu, Mario Bauer, Matthias Bieg, Zuguang Gu, Dieter Weichenhan, Jan‐Philipp Mallm, Stefan Röder, Gunda Herberth, Eiko Takada, Oliver Mücke, Marcus Winter, Kristin M Junge, Konrad Grützmann, Ulrike Rolle‐Kampczyk, Qi Wang, Christian Lawerenz, Michael Borte, Tobias Polte, Matthias Schlesner, Michaela Schanne, Stefan Wiemann, Christina Geörg, Hendrik G Stunnenberg, Christoph Plass, Karsten Rippe, Junichiro Mizuguchi, Carl Herrmann, Roland Eils, Irina Lehmann
Molecular Systems Biology (2016)
Increased vitamin D levels at birth and in early infancy increase offspring allergy risk–evidence for involvement of epigenetic mechanisms
Kristin M Junge, Tobias Bauer, Stefanie Geissler, Frank Hirche, Loreen Thürmann, Mario Bauer, Saskia Trump, Matthias Bieg, Dieter Weichenhan, Lei Gu, Jan-Philipp Mallm, Naveed Ishaque, Oliver Mücke, Stefan Röder, Gunda Herberth, Ulrike Diez, Michael Borte, Karsten Rippe, Christoph Plass, Carl Hermann, Gabriele I Stangl, Roland Eils, Irina Lehmann
Journal of Allergy and Clinical Immunology (2016)
Evolutionary conserved gene co-expression drives generation of self-antigen diversity in medullary thymic epithelial cells
Kristin Rattay, Hannah Verena Meyer, Carl Herrmann, Benedikt Brors, Bruno Kyewski
Journal of autoimmunity (2016)
Telomerase activation by genomic rearrangements in high-risk neuroblastoma
Martin Peifer, Falk Hertwig, Frederik Roels, Daniel Dreidax, Moritz Gartlgruber, Roopika Menon, Andrea Krämer, Justin L Roncaioli, Frederik Sand, Johannes M Heuckmann, Fakhera Ikram, Rene Schmidt, Sandra Ackermann, Anne Engesser, Yvonne Kahlert, Wenzel Vogel, Janine Altmüller, Peter Nürnberg, Jean Thierry-Mieg, Danielle Thierry-Mieg, Aruljothi Mariappan, Stefanie Heynck, Erika Mariotti, Kai-Oliver Henrich, Christian Gloeckner, Graziella Bosco, Ivo Leuschner, Michal R Schweiger, Larissa Savelyeva, Simon C Watkins, Chunxuan Shao, Emma Bell, Thomas Höfer, Viktor Achter, Ulrich Lang, Jessica Theissen, Ruth Volland, Maral Saadati, Angelika Eggert, Bram De Wilde, Frank Berthold, Zhiyu Peng, Chen Zhao, Leming Shi, Monika Ortmann, Reinhard Büttner, Sven Perner, Barbara Hero, Alexander Schramm, Johannes H Schulte, Carl Herrmann, Roderick J O’Sullivan, Frank Westermann, Roman K Thomas, Matthias Fischer
Nature (2015)
MYCN-driven regulatory mechanisms controlling LIN28B in neuroblastoma
Anneleen Beckers, Gert Van Peer, Daniel R Carter, Moritz Gartlgruber, Carl Herrmann, Saurabh Agarwal, Hetty H Helsmoortel, Kristina Althoff, Jan J Molenaar, Belamy B Cheung, Johannes H Schulte, Yves Benoit, Jason M Shohet, Frank Westermann, Glenn M Marshall, Jo Vandesompele, Katleen De Preter, Frank Speleman
Cancer letters (2015)
MapMyFlu: visualizing spatio-temporal relationships between related influenza sequences
Nicholas Nolte, Nils Kurzawa, Roland Eils, Carl Herrmann
Nucleic acids research (2015)
RSAT 2015: regulatory sequence analysis tools
Charles E Chapple, Carl Herrmann, Christine Brun
Front. Genet. (2015).
Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state
Annelien Verfaillie, Hana Imrichova, Zeynep Kalender Atak, Michael Dewaele, Florian Rambow, Gert Hulselmans, Valerie Christiaens, Dmitry Svetlichnyy, Flavie Luciani, Laura Van den Mooter, Sofie Claerhout, Mark Fiers, Fabrice Journe, Ghanem-Elias Ghanem, Carl Herrmann, Georg Halder, Jean-Christophe Marine, Stein Aerts
Nature Communications (2015).