[BC]2 Tutorial
Defining genomic signatures with Non-Negative Matrix Factorization

Carl Herrmann & Andres Quintero
13 September 2021
ButcherR-[BC]2 Docker image
We created a Docker image that runs RStudio, including all the package dependencies, and example datasets required to run a complete NMF workflow and infer molecular signatures.
Step 1 - Pull image
- Open the Docker application in your computer
- Open a command-line terminal (e.g., Command Promt and Powershell in windows, or Terminal in Max and Linux).
- Pull the image by running the following command:
docker pull hdsu/butchr-bc2
IMPORTANT NOTE!
The image size is approximately 6Gb. Therefore, we ask you to complete this step before starting the tutorial.
Step 2 - Running the image
- Once the image has been pulled from DockerHub you can run it from the command-line, using the following command:
docker run --rm -p 8787:8787 -e USER=hdsu -e PASSWORD=pass hdsu/butchr-bc2
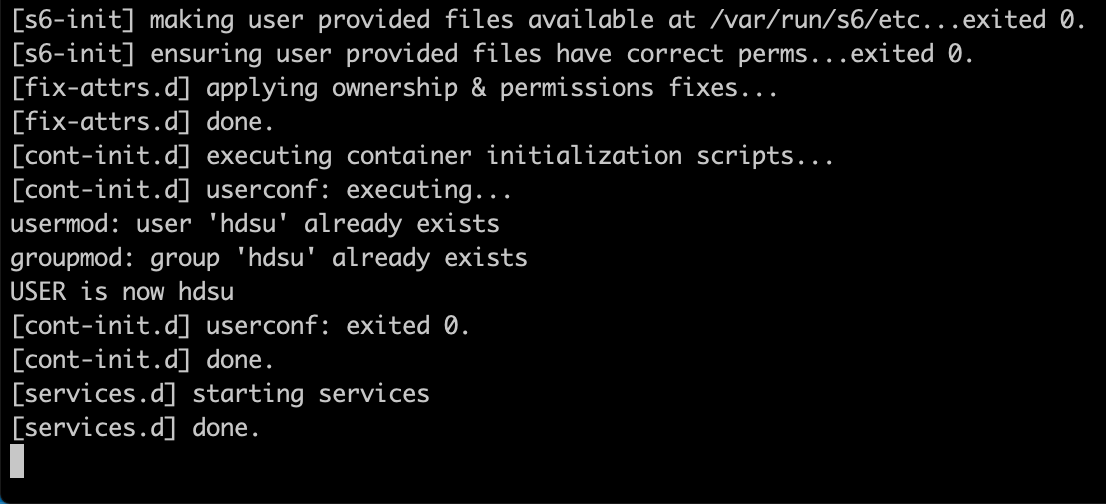 |
|---|
| Output messages expected after running the previous command |
- Open the app in a browser: http://localhost:8787/
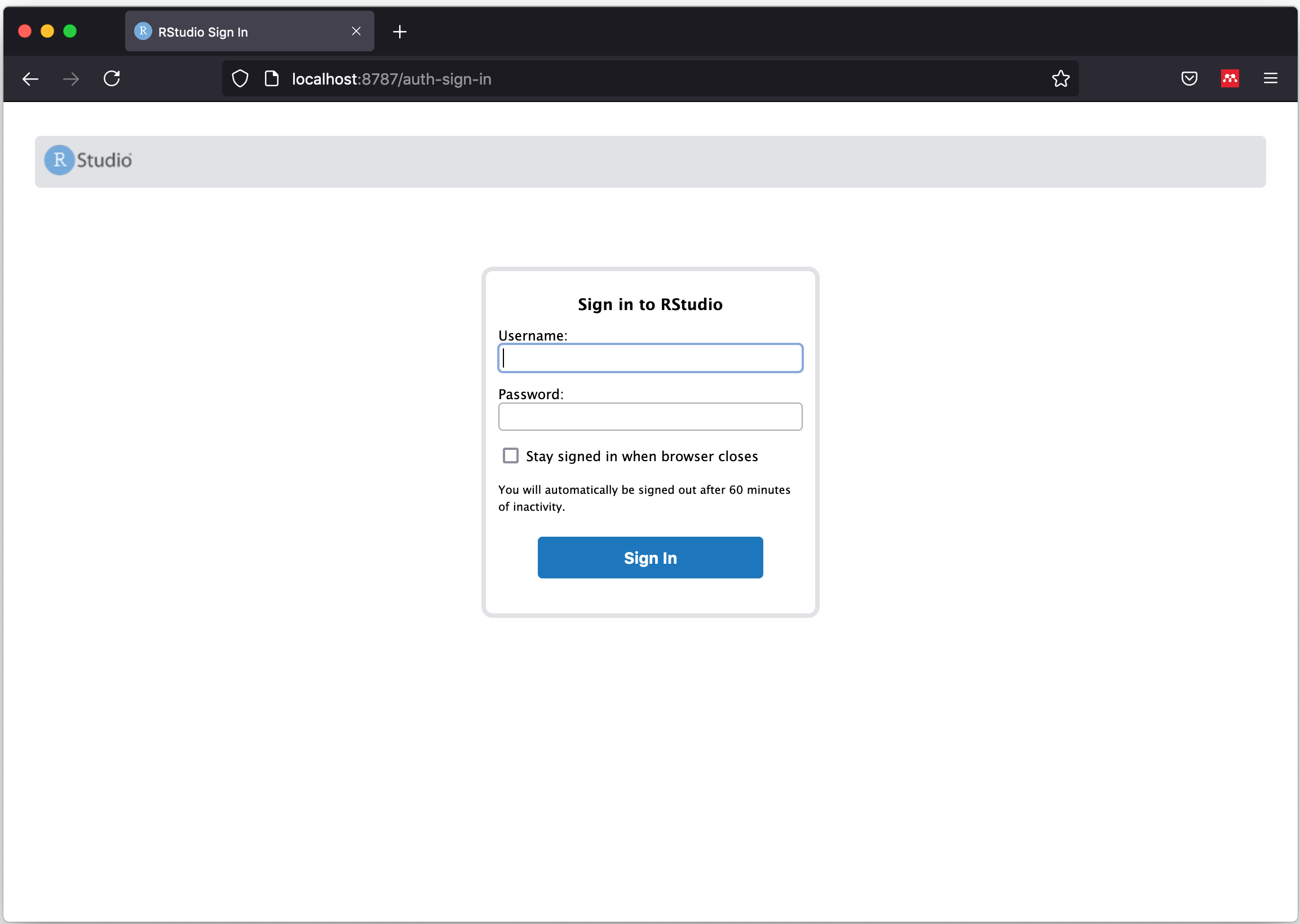 |
|---|
| First screen seen after launching the image without errors |