IRTG Course
Introduction to R for genomics
Carl Herrmann & Carlos Ramirez
8-9 December 2021Dimensional Reduction
The size of scRNA-Seq matrices can be huge and for this reason techniques to reduce the dimensionality of this data are used. Here, we will use PCA, a very common techniques for dimension reduction and visualization.
We will run a PCA using the already calculated top 1000 HVGs using the function RunPCA().
pbmc.filtered <- RunPCA(pbmc.filtered,
features = VariableFeatures(pbmc.filtered))
We can assess the dimensionality, a measure of the complexity, by using an elbow plot of the standard deviation for each principal component (PC) from the PCA.
We will use the function ElbowPlot().
ElbowPlot(pbmc.filtered)
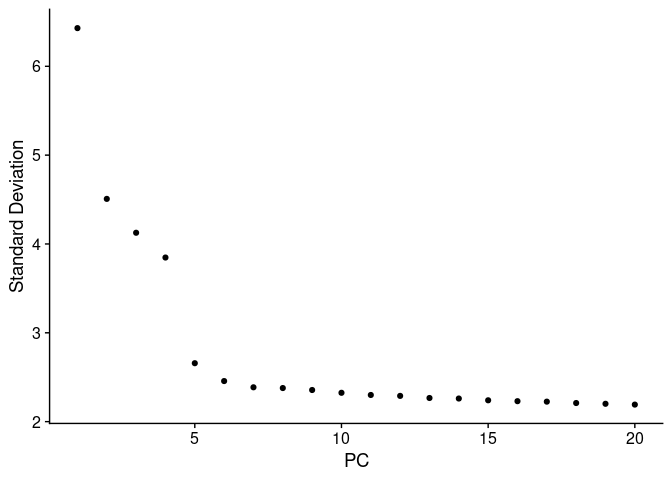
The PC components in a PCA reflects corresponds to the directions in which more variability is observed. These PCs are ranked by using the eigenvalues of the covariance matrix. We can the plot a Elbow or joystick plot of the standard deviation and the rank of each PC. Top ranked PCs are expected to have higher values of variability and then to gradually decrease. So, we can use the elbow plot representation to keep PCs from the top to the bottom until we do not see further variability changes, in these case we can use the number of PC equal to 7.
Quizzes
Manipulation of matrix of PCA
Load a seurat object using the following command:
pbmc.seurat <- readRDS(url('https://raw.githubusercontent.com/caramirezal/caramirezal.github.io/master/bookdown-minimal/data/pbmc_10X_250_cells.seu.rds'))
Perform the seurat standard pre-processing pipeline from the previous sections. Extract PCA embedding matrix and make a PCA plot showing the first 2 principal components.
pca <- Embeddings(pbmc.seurat, reduction = 'pca')pca <- pbmc.seurat@reductions$pca@cell.embeddingspca <- pbmc.seurat@pca$reductions@cell.embeddings
TIP: Use str(pbmc.seurat) to explore the slots present in the seurat object. Two options are correct.