Workshop ChIPATAC 2020
Computational analysis of ChIP-seq and ATAC-seq data
14-15 December 20201. ChIP-seq : Introduction and data
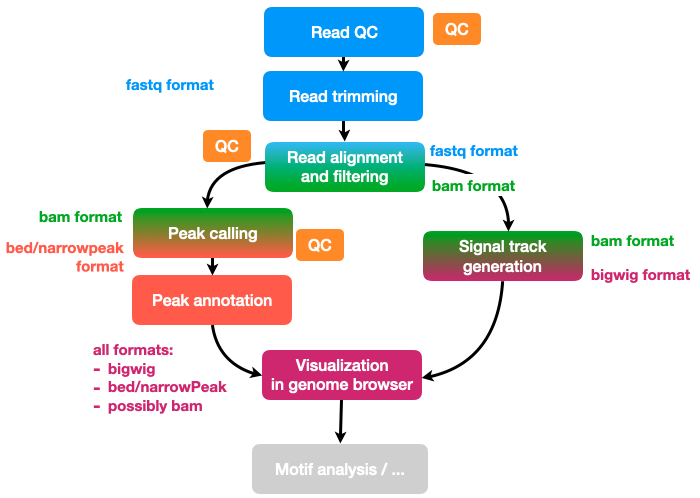
Data background
In this workshop, we will analyse different genomic data generated from wild-type HCT116 colon cancer cell-lines. These data will comprise of genome-wide chromatin accessibility measurements using ATACseq, histone modification of H3K4me3 marks and CTCF transcription factor binding using ChIPseq. We also downloaded the H3K27ac data, which you can use to reproduce the whole analysis from scratch.
H3K4me3 is known to mark active promoters around their TSS, H3K4me1 are known to mark enhancer regions and the transcription factor CTCF is known to mediate enhancer-promoter interactions. Also, genomic regions of activity (active promoter or enhancers) are expected to have an open chromatin landscape to allow the binding of regulatory factors. Thus, integration of ATACseq data and ChIPseq data (H3K4me3, H3K27ac and CTCF) from the same cell-line will give us an understanding of its regulatory landscape.
Raw data will be downloaded and re-processed from ENCODE and the following data will be analyzed -
| Sample | Experiment | Assay | Replicate | Source |
|---|---|---|---|---|
| HCT116 | ChIPseq | Histone modification - H3K4me3 | 1 | ENCFF001FIS |
| HCT116 | ChIPseq | Histone modification - H3K4me3 | 2 | ENCFF001FIZ |
| HCT116 | ChIPseq | ChIPseq control - H3K4me3 | 1 | ENCFF001HME |
| HCT116 | ChIPseq | Histone modification - H3K27ac | 1 | ENCSR661KMA |
| HCT116 | ChIPseq | ChIPseq control - H3K27ac | 1 | ENCSR198WIH |
| HCT116 | ChIPseq | Transcription factor - CTCF | 1 | ENCFF001HLV |
| HCT116 | ChIPseq | Transcription factor - CTCF | 2 | ENCFF001HLW |
| HCT116 | ChIPseq | ChIPseq control - CTCF | 1 | ENCFF001HME |
Download data
NOTE: You don't need to download any data, all of the data has been pre-downloaded and saved in
/home/<username>/data/fastqdata. You will directly access these data during the practical training session.
ChIPseq - H3K4me3
We have downloaded the fastq files (2 isogenic replicates and 1 control) containing sequence reads (single end) from a H3K4me3 ChIPseq experiment done on the HCT116 cell-line from the ENCODE database.
wget https://www.encodeproject.org/files/ENCFF001FIS/@@download/ENCFF001FIS.fastq.gz \
-O H3K4me3_Rep1_ENCFF001FIS.fastq.gz ;
wget https://www.encodeproject.org/files/ENCFF001FIZ/@@download/ENCFF001FIZ.fastq.gz \
-O H3K4me3_Rep2_ENCFF001FIZ.fastq.gz ;
wget https://www.encodeproject.org/files/ENCFF001HME/@@download/ENCFF001HME.fastq.gz \
-O H3K4me3_Control_ENCFF001HME.fastq.gz
ChIPseq - CTCF
We have downloaded the fastq files (2 isogenic replicates and 1 control) containing sequence reads (single end) from a CTCF ChIPseq experiment done on the HCT116 cell-line from the ENCODE database.
wget https://www.encodeproject.org/files/ENCFF001HLV/@@download/ENCFF001HLV.fastq.gz \
-O CTCF_Rep1_ENCFF001HLV.fastq.gz ;
wget https://www.encodeproject.org/files/ENCFF001HLW/@@download/ENCFF001HLW.fastq.gz \
-O CTCF_Rep2_ENCFF001HLW.fastq.gz ;
wget https://www.encodeproject.org/files/ENCFF001HME/@@download/ENCFF001HME.fastq.gz \
-O CTCF_Control_ENCFF001HME.fastq.gz
hg38 genome index
We have also downloaded the precomputed hg38 genome index reference for alignment from iGenomes
wget http://igenomes.illumina.com.s3-website-us-east-1.amazonaws.com/Homo_sapiens/NCBI/GRCh38/Homo_sapiens_NCBI_GRCh38.tar.gz
Public datasets
Often one has to use publicly available datasets. These datasets are widely available through Gene Expression Omnibus - GEO and Array Express. Raw data fastq from human samples are usually deposited in The database of Genotypes and Phenotypes - dbGAP and European Genome-phenome Archive - EGA and are available under protected access. These data are usually in SRA format and an array of tools called sra-tools are available to manipulate these formats prior to regular analysis. We will not talk about these files formats in this workshop, but we want to make the participants aware that most of the publicly available raw ChIPseq and ATACseq data are in SRA format which needs to be converted to fastq format using SRA-tools.